This project supports development for two lab-based quantitative molecular methods for more rapid, accurate detection of Alexandrium catenella resting cysts in sediment from the Gulf of Maine, Puget Sound, and the waters around Kodiak Island and Kachemak Bay, AK. Winter abundance of benthic Alexandrium cysts is used to predict severity of the spring bloom but cyst enumeration is time and labor intensive. Molecular detection methods and training is needed to advance research and forecasting applications that reduce effort and turnaround time for cyst enumeration. The project will expand NOAA HAB Operational Forecasting to new regions.
Why We Care
Along our Northeast and Pacific Northwest coasts, annual Alexandrium catenella blooms can produce potent neurotoxins that accumulate in shellfish and cause Paralytic Shellfish Poisoning (PSP), a potentially fatal illness affecting shellfish consumers. NOAA’s Harmful Algal Bloom (HAB) Operational Forecasting System is used to produce forecast products for toxic Alexandrium blooms. These products help coastal states mitigate human health risks from PSP and economic effects of shellfish closures.
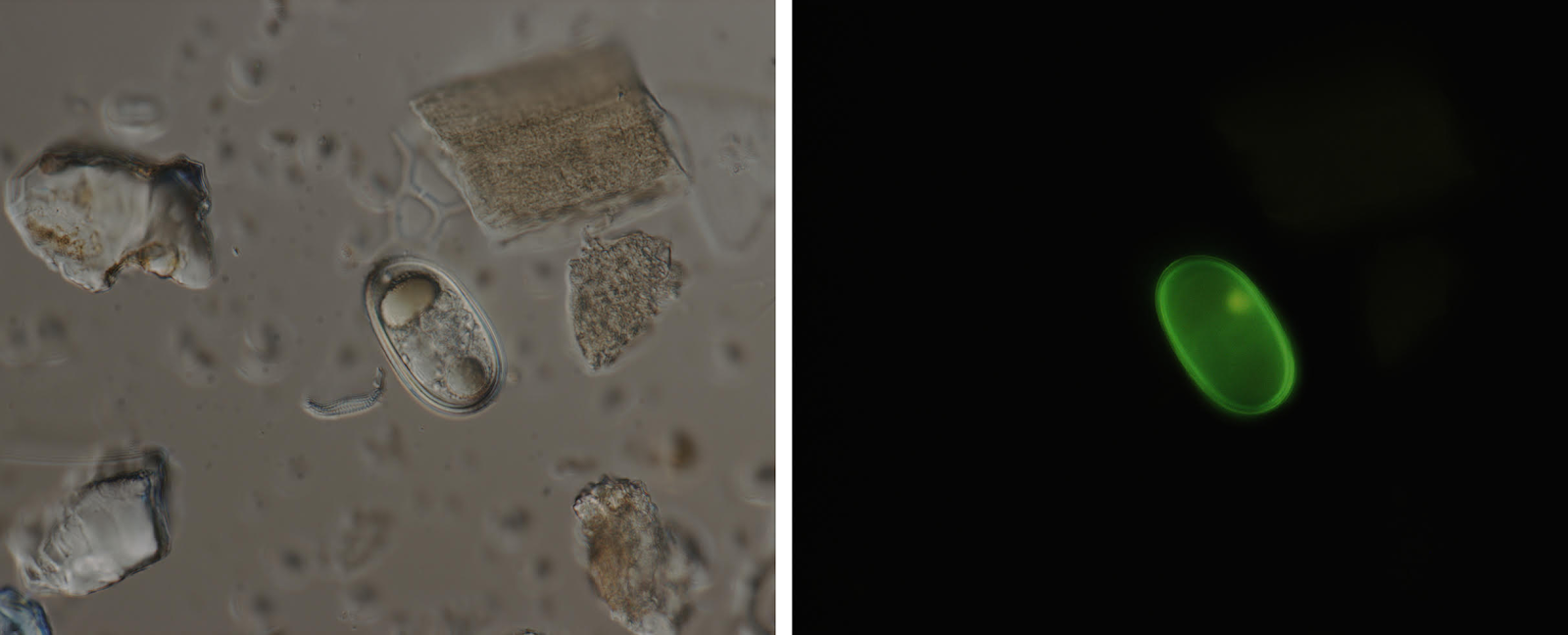
Forecasting efforts hinge on determining winter abundance of benthic Alexandrium resting cysts, which germinate to initiate blooms each spring. Building on demonstrated success in the Gulf of Maine, NOAA and partners are developing HAB forecasting products for Puget Sound and central Alaska to help mitigate PSP risks in these regions. The current protocol for cyst enumeration by fluorescent microscopy is laborious, requires highly specific training, and has some uncertainty about the identity of A. catenella resting cysts and their vitality. There is a critical need to reduce the time and effort required to produce the Alexandrium cyst data for these forecast before the spring bloom season.

What We Are Doing
This project advances two species-specific molecular techniques developed previously for vegetative cells using a quantitative polymerase chain reaction (qPCR)-based method and a fluorescent in situ hybridization (FISH)-based method. The project team will design qPCR and FISH assays using A. catenella resting cysts in sediment samples collected in 2018 from the three study locations. New samples collected after the 2019 and 2020 summer bloom seasons will allow for direct comparison of microscopy-based and molecular assay-based cyst abundance data. As a substitute for the cyst sample processing, staining and microscopy currently used, these two molecular based analyses can be completed in a fraction of the time and cost (1-2 days of manpower resources).
Additional samples from collaborators on an ECOHAB project in Southeast Alaska will make the comparison more robust. After validation, the new methods will be shared with other HAB researchers, monitoring organizations and stakeholders in the Gulf of Maine, the State of Washington, Alaska and Canada through public presentations, publications, and a training workshop at the 2021 U.S. National HAB Meeting. The resulting new molecular tools will also be integrated into the NOAA HAB Operational Forecasting System to enable streamlined, less costly generation of critical cyst abundance data. The study results will be published in an open access scientific journal. If application of the new methods is successful, a demonstration workshop will be organized during the 11th US Symposium on Harmful Algae in the fall of 2021.
Dr. Cheryl Greengrove of University of Washington-Tacoma leads this project. Co-investigators are Julie Masura (University of Washington-Tacoma), Julie Matweyou (University of Alaska Fairbanks, Alaska Sea Grant), Steve Kibler (NOAA NCCOS) and will involve undergraduate and graduate students.
Benefits Of This Work
The project, funded through the NCCOS Monitoring and Event Response Harmful Algal Blooms (MERHAB) Program, will help NOAA and its regional partners address MERHAB priorities to mitigate HAB impacts by improving existing methods of monitoring, comparing new and existing monitoring systems, and helping to develop forecasting technologies.


